- Academic Editor
†These authors contributed equally.
Background: The antibiotic resistance of microorganisms is escalating rapidly. Infections caused by opportunistic pathogens in immunocompromised individuals have prompted researchers to seek for potent and safe antibacterial agents. The purpose of this investigation was to explore the suppression of virulence gene expression, specifically the pga operon genes responsible in biofilm formation in Acinetobacter baumannii, through the utilization of metabolites obtained from probiotic bacteria. Methods: To assess the antimicrobial properties, standard strains of five probiotic bacteria were tested against a standard strain of multidrug-resistant (MDR) A. baumannii employing the agar gel diffusion technique. Following the identification of the most potent probiotic strain (Bacillus licheniformis), the existence of its LanA and LanM genes was confirmed using the polymerase chain reaction (PCR) test. High-performance liquid chromatography (HPLC) and fourier-transform infrared spectroscopy (FTIR) techniques were employed to identify the intended metabolite, which was found to be a lipopeptide nature. The minimum inhibitory concentration (MIC) values and anti-biofilm activity of the targeted metabolite were determined using a dilution method in 96-well microplates and field emission scanning electron microscopy (FE-SEM). Real-time PCR (qPCR) was utilized for comparing the expression of pga operon genes, including pgaABCD, in A. baumannii pre- and post-exposure to the derived lipopeptide. Results: The MIC results indicated that the probiotic product inhibited the growth of A. baumannii at concentrations lower than those needed for conventional antibiotics. Furthermore, it was observed that the desired genes’ expression decreased due to the effect of this substance. Conclusions: This research concludes that the B. licheniformis probiotic product could be a viable alternative for combating drug resistance in A. baumannii.
Recently, opportunistic infections have created multiple problems in managing the treatment of patients [1]. Nosocomial infections are a common and important cause of death, disability, Long-term hospitalization, increasing costs, and occurrence of health risks [2]. These infections threaten almost all hospitalized patients, and can lead to patient death [3]. Treating these infections proves to be notably tough as a consequence of the antibiotic resistance found in the majority of microbial isolates, and it is expensive due to the long hospitalization time of the patients [4]. Among the opportunistic pathogens found in the hospital environment, particularly in the intensive care unit (ICU), A. baumannii is important due to the occurrence of various complications such as burn infections, wounds, endocarditis, bacteremia, meningitis, and hospital-acquired pneumonia [5, 6]. The effective factors in the virulence of A. baumannii are the presence of lipopolysaccharide, outer membrane proteins, stress response mechanisms, and biofilm formation [7]. Quorum sensing (QS) is another factor that contributes to the virulence and pathogenicity of A. baumannii, particularly in relation to biofilm formation. In general, there are at least two main types of QS, the first type is involved in intra-species communication and the second type is involved in inter-species communication [8, 9]. The roles of QC include cell division (production of exoenzymes and siderophores), gene transfer, host-pathogen interaction, and behavior (movement and biofilm formation) [10]. A. baumannii has garnered interest in recent times for its inherent resistance to a wide range of antibiotics [11]. Among the features that contribute to this resistance, we can highlight the ability to acquire antibiotic resistance genes, presence of efflux pumps, multiple beta-lactamase enzymes, and the capacity to form a robust biofilm [12]. A biofilm is a formation in which clusters of microorganisms are enveloped by exopolysaccharides produced by the microbes themselves, enabling them to adhere to diverse surfaces and hosts [13]. The biofilm structure consists of proteins, polysaccharides, DNA, and RNA. Moreover, water constitutes a significant portion, accounting for 97% of the biofilm. This water component plays a vital role in facilitating the transport of nutrients within the biofilm matrix. Furthermore, microorganisms residing within the biofilm find protection from microbial competition, fluctuating temperatures, host cells, antimicrobial substances, and desiccation [9].
The complex matrix of biofilms is composed of protein, nucleic acid, and polysaccharide polymers [14]. Exopolysaccharides play a superglue role in biofilm formation and are complex combinations of biopolymers containing polysaccharides, proteins, DNA, and phospholipids, which these contents depend on the type of pathogens, age of biofilms and environmental conditions (dryness, pH, levels of oxygen and nitrogen, temperature, and available nutrients) can be changed [15]. Poly-beta-1–6-N-acetylglucosamine (PNAG), a crucial polysaccharide, is synthesized in A. baumannii by the pga operon, consisting of the pgaA, pgaB, pgaC, and pgaD genes [16]. This genetic operon is important because it encodes proteins that are responsible for the synthesis of an extracellular polysaccharide called PNAG [17]. This polymer is the agent of connecting bacterial cells to each other and surfaces, as well as an important factor of virulence and protection of bacteria against the host’s innate defense [18]. PNAG is the main component of the biofilm matrix in A. baumannii and its synthesis is vital for the formation of biofilm, the accumulation of bacteria and their protection [19]. PNAG promotes the biofilm integrity and enhanced the bacterial tolerance to desiccation stress in natural environments and medical settings [20]. Under stressful conditions, transcription of the pga operon genes increases and leads to the formation of thicker cell walls against this stress (environmental drought); therefore, this operon plays a vital role in the stability of A. baumannii on non-living surfaces [21]. By deleting the pga operon, researchers observed that A. baumannii lacked PNAG and the bacterium lost its capability to form a robust biofilm [22, 23].
Probiotics, consisting of bacteria and yeast, typically promote a healthy equilibrium of the body’s natural flora [24]. Generally, probiotics are alive microbial dietary supplements that can be incorporated into different types of foods, pharmaceuticals, and other products [25]. Probiotics provide consumers with health benefits when consumed in appropriate amounts [26]. It is essential for probiotics to possess non-pathogenic characteristics and fall under the category of GRAS (Generally Recognized as Safe) microorganisms [27].
Antimicrobial chemicals are extensively employed to combat various infections [28]. However, their effectiveness is currently hindered by the rise of antibiotic-resistant strains and associated side effects [29]. Consequently, researchers are constantly seeking potent and safe antimicrobial agents. In addressing this issue, probiotics have proven to be highly successful [27]. One of the traits exhibited by probiotics is the presence of LanA and LanM lantibiotic genes within bacteria [30]. Lantibiotics are antibiotic compounds that are produced by microorganisms in the form of small ribosomal peptides [31]. These antibiotics belong to the group of bacteriocins, have low molecular weights, and their mechanism of antibacterial action is to disrupt cell membranes or stop the activity of microbial cellular enzymes [32].
Different Bacillus and Lactobacillus strains produce peptide antibiotics such as gramicidins, bacitracins, tyrocidines, and different biological such as lipopeptides, proteases, antibiotics, bacteriocins, surfactants, and various compounds that have minimal risks to human health. A variety of beneficial microbial metabolites can be produced, including these compounds [33]. Some of these biomaterials possess antibacterial properties and compared to other antibacterial materials, lipopeptides are more environmentally friendly and have a little environmental impact [34].
Lipopeptides are compounds with a cyclic peptide linked to a long fatty acid chain, and they have a low molecular weight and amphiphilic nature. Bacterial strains generally produce lipopeptides via the ribosomal or non-ribosomal pathway. In bacilli, lipopeptides are synthesized in a ribosome-independent manner by non-ribosomal peptide synthetases [35]. Gram-positive bacteria, especially bacilli, have the ability to generate a diverse range of antimicrobial cyclic lipopeptides (consisting of 98 distinct lipopeptide types), whereas Gram-negative bacteria produce rhamnolipids and glycolipid biosurfactants [36].
The aim of this research was to examine the impact of metabolites derived from probiotic bacteria on the expression of pgaA, pgaB, pgaC, and pgaD virulence genes, which are associated with biofilm formation in multidrug-resistant (MDR) A. baumannii.
This study aimed to examine the antibacterial characteristics of five commonly used probiotic bacteria strains, namely Lactobacillus rhamnosus American Type Culture Collection (ATCC) 52103, Pediococcus acidilactici ATCC 8042, Bacillus coagulans ATCC 31284, Bacillus subtilis ATCC 21335, and B. licheniformis ATCC 21415, against the reference strain of MDR A. baumannii ATCC BAA-747. These reference strains were developed by the Microbiological Standardization Department of the Pasteur Institute in Iran.
Lactobacilli deMan, Rogosa, and Sharpe (MRS) media is an adapted version of MRS formulation with slight modification. For each strain, 50 mL of MRS broth medium (Merck, Darmstadt, Germany) was prepared and incubated at 37 °C for 48 hours after bacterial inoculation. After 48 hours, the bacterial suspension was centrifuged at 7000 rpm for 15 minutes at 4 °C, and the supernatant was collected [37].
For each strain, 100 mL of Luria broth medium (Merck) was prepared. Each strain was separately inoculated into Erlenmeyer flasks and incubated at 30 °C for 48 hours in a shaking incubator set at 175 rpm. After 48 hours, the bacterial suspension was transferred to Falcon tubes, centrifuged at 10,000 rpm for 10 minutes at 4 °C, and the supernatant was collected [38].
The Kirby-Bauer method was employed to evaluate the susceptibility of 20 different antibiotic disks, in accordance with the guidelines established by the Clinical and Laboratory Standards Institute (CLSI). This study included different classes of antibiotics, such as beta-lactams, aminoglycosides, tetracyclines, and macrolides [39].
The antibiotics utilized in the study included ceftriaxone (30 µg), ceftazidime (30 µg), gentamicin (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), piperacillin (100 µg), piperacillin-tazobactam (100/10 µg), imipenem (10 µg), meropenem (10 µg), aztreonam (30 µg), cefepime (30 µg), cefotaxime (5 µg), tobramycin (10 µg), ticarcillin-clavulanic acid (75 µg), polymyxin B (75 µg), ampicillin-sulbactam (20 µg), levofloxacin (5 µg), cotrimoxazole (40 µg), doxycycline (40 µg), and colistin (10 µg). These antibiotics were sourced from MAST Co. (Merseyside, UK).
To assess the sensitivity or resistance of each antibiotic using the Kirby-Bauer method, antibiotic discs were positioned at specific intervals on a Mueller Hinton Agar (MHA) (Merck, Darmstadt, Germany) culture medium plate. Once the bacteria had adequately multiplied, the diameter of the zone of growth inhibition beneath the antibiotic discs was evaluated. The interpretation of the results was then done based on the CLSI breakpoints [40].
After collecting the supernatant from each probiotic strain, we examined the antimicrobial characteristics of these strains using the agar well diffusion technique. To perform this method, a fresh culture suspension of the A. baumannii standard strain ATCC BAA-747 was prepared at a half McFarland dilution. The suspension was then cultured on the MHA plate using a sterile swab. Then, using a Pasteur pipette, 6 mm wells were created on the plate. 50 µL of the supernatant from each strain of probiotics were individually transferred into each well. The metabolite obtained from B. licheniformis showcased an outstanding antibacterial impact, surpassing that of other probiotics.
The presence of two genes, lanA and lanM, in B. licheniformis was determined through polymerase chain reaction (PCR) technology. To achieve this, the genome was extracted by the boiling method and added to a final volume of 20 µL for the reaction. A Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) at A260/A280 nm was used to quantify the extracted genome. In our work, the ratio of 260/280 was 1.85, which indicated that the DNA had a good purity [41]. The reaction mixture consisted of 1 µL each of forward and reverse primers (Table 1), 12.5 µL of master mix, 1 µL of the extracted genome, and 4.5 µL of sterile distilled water.
| LanM-For | AAGCGGCTGAGCATTTGG | 287 bp | [This study] |
| LanM-Rev | GTCGGCGTACTTCAGGAATTC | ||
| LanA-For | CCCGCAAATCATTAATCAGC | 724 bp | [This study] |
| LanA-Rev | GACGGCGTTCTTAAATGGG |
The thermal program employed in the PCR reaction involved an initial denaturation step at 94 °C for 4 minutes, followed by denaturation at 94 °C for 1 minute. The annealing temperature for the specific primer targeting the lanA gene was set at 53 °C, while the annealing temperature for the specific primer targeting the lanM gene was set at 52 °C, both for a duration of 1 minute. The amplification step occurred at 72 °C for 30 seconds, repeated for a total of 30 cycles. Lastly, there was a final amplification step at 72 °C for 5 minutes. Subsequently, the PCR product was evaluated using a 1% agarose gel analysis [42].
The supernatant extracted from B. licheniformis was mixed with ethyl acetate in a decanter flask, keeping a 1:1 volume ratio (V/V), and the upper phase was filtered through a 0.45 µm filter. Next, the ethyl acetate was dried using a rotary evaporator (Boeco, Hamburg, Germany). Then, high-performance liquid chromatography (HPLC) and fourier-transform infrared spectroscopy (FTIR) methods were utilized to analyze and determine the structure [36]. The desired lipopeptide was assessed for its minimum inhibitory concentration (MIC) by preparing an initial concentration of 32 µg/mL from the resulting powder of the purified probiotic product, which was powdered using the Zirbus freeze-dryer (ZIRBUS technology GmbH, Bad Grund, Germany).
HPLC enables the separation of compound mixtures and can be utilized for identifying, quantifying, and purifying various compounds. In this study, the HPLC device used was the KNAUER model D-14163 from Berlin, Germany. The mobile phase consisted of 1% acetic acid and acetonitrile in a ratio of 60:40 (V/V), while the stationary phase employed was the C18 column. This system incorporates a Maxi-star K-1000 model pump in conjunction with a UV-Vis spectrophotometer detector (K-2500 model), which was configured to function at a wavelength of 280 nm. For this experiment, a mobile phase comprising a blend of 1% acetic acid and acetonitrile with a 60:40 (V/V) ratio was employed, utilizing the isocratic method at a flow rate of one milliliter per minute. The column utilized was C18 phase with dimensions of 25 cm in length, 4.6 mm in diameter, and containing 5 µm ODS-silica particles. A sample volume of 20 µL, with a concentration of 100 µg/L in the mobile phase, was injected. The quantitative identification and measurement were performed utilizing a calibration curve and by comparing the retention time (RT) with the international standard [43].
This method provides a rapid and cost-effective means of describing chemical
structures and identifying functional groups within compounds. The technique
involves measuring the molecular vibrations induced by specific infrared
radiation wavelengths. To conduct this analysis, dry metabolite powder obtained
from the supernatant of B. licheniformis was utilized. The samples were
examined using KBr cells for analysis. The FTIR analysis was performed using
Jasco (Tokyo, Japan) spectrometer, and the samples were evaluated within the wavelength
range of 4000 to 400 cm
For this purpose, a germ suspension was prepared from a logarithmically growing
standard strain of A. baumannii with a concentration of 10
To begin, a concentration of 10
For staining, 200 µL of a 1% crystal violet solution was added to the
wells and allowed to sit for 15 minutes. Following this, the dye was removed; the
wells were rinsed three times with physiological serum, and then dried once more
at room temperature. Next, 200 µL of 95% ethanol was added to the wells.
The absorbance of each well was measured at a wavelength of 578 nm using Eliza
Reeder (Biotech, Winooski, VT, USA). Furthermore, we measured the state of biofilm formation in
each well by examining the optical absorbance (OD) and the optical absorbance
control (ODC), so that if OD
A microbial suspension was prepared from logarithmic growth phase standard
strains of A. baumannii, with a concentration of 10
The genome of the standard strain of A. baumannii was extracted using the boiling method and transferred into a microtube, which was then stored at a temperature of –20 °C. The PCR reaction consisted of 1 µL of forward and 1 µL of reverse primer (Table 2), 12.5 µL of master mix (dNTP, Mgcl2, and a Taq DNA polymerase enzyme), 1 µL of the extracted genome of A. baumannii strain, and 4.5 µL of sterile distilled water in a final volume of 20 µL.
| Names of genes | Primer sequence (5 |
PCR product size | Reference |
| pgaA-For | CAGCAAATGAATCCTTCCGATCCT | 605 bp | This study |
| pgaA-Rev | GCGGGTGTCTTTAGCAATCGGATC | ||
| pgaB-For | GAGGGTATGATCAATATTCTCAAC | 400 bp | This study |
| pgaB-Rev | CCAGCATTAAGTTGAATACCACTT | ||
| pgaC-For | GGTTCGCGTGGGTTTTGGTCTGATGA | 518 bp | This study |
| pgaC-Rev | CACTGGTAAATAAGGTCAGTAACC | ||
| pgaD-For | GACTATGTGTGGGTCGATCATGTT | 224 bp | This study |
| pgaD-Rev | GCGCTGGGAATTTTGTAATTCTGA |
The thermal program utilized in the PCR involved an initial denaturation cycle at 95 °C for 5 minutes, followed by 30 cycles. Each cycle consisted of denaturation at 94 °C for 1 minute. Specific primer binding occurred at different temperatures: 53 °C for the pgaA gene, 57 °C for the pgaB gene, 54 °C for the pgaC gene, and 52 °C for the pgaD gene, each for 1 minute. Subsequently, there was an amplification step at 72 °C for 1 minute specifically targeting the pga operon genes. The process was concluded with a final amplification phase at 72 °C for 5 minutes. A holding temperature of 20 °C was maintained throughout the procedure [42].
The expression of pga operon genes, involved in biofilm formation in MDR A. baumannii, was investigated using this method. Specifically, a relative qPCR test was conducted to compare the expression of genes before and after 24-hour treatment with antibiotics and metabolite derived from the supernatant of B. licheniformis. For RNA extraction and cDNA synthesis, a kit from Biofact (Celbridge, Ireland) was employed.
Following RNA extraction and cDNA synthesis, the desired gene expression was examined using the qPCR technique. The reaction mixture comprised 12.5 µL of 2X Sybergreen master mix, 1 µL of each forward & reverse primers (Table 3, Ref. [49, 50]), and 5.5 µL of cDNA, resulting in a final volume of 20 µL. The temperature program for the reaction involved an initial denaturation step at 95 °C for 4 minutes, denaturation at 94 °C for 10 seconds, primer binding at 58 °C for 20 seconds, and extension at 72 °C for 25 seconds. This cycle was repeated 30 times.
| Names of genes | Primer sequence (5 |
Reference |
| pgaA-For | CCCGCTTCAAAATGCTGCTT | [49] |
| pgaA-Rev | AAGGCTATTCGACGCACCTC | |
| pgaB-For | ATGCATCTGGGCTTGCATCT | [49] |
| pgaB-Rev | GTGACAGAGCAGGCAAAAGC | |
| pgaC-For | ACGAATCGTTTGTCGGACCT | [49] |
| pgaC-Rev | ATGGTAGGAGGCCTCTGGTT | |
| pgaD-For | GTGGCTGCTTTTTCCACTCG | [49] |
| PgaD-Rev | CGCGCTCAGACCTATTAGCA | |
| 16sDNA-For | GTAATACAGAGGGTGCGAGCGTT | [50] |
| 16sDNA-Rev | TCTAGCTGACCAGTATCGAATGCA |
The antimicrobial property of the supernatant obtained from strains of L. rhamnosus, P. acidilactici, B. subtilis, B. coagulans, and B. licheniformis against the MDR standard strain of A. baumannii was examined utilizing the agar well diffusion technique. Among the probiotic strains tested, only the B. licheniformis strain demonstrated the ability to inhibit the growth of antibiotic-resistant strain, resulting in a non-growth zone including a well with a diameter of 10 mm (Fig. 1).
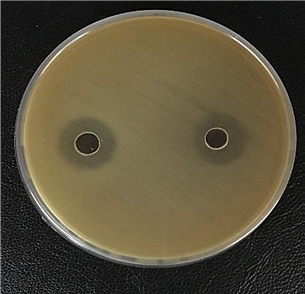 Fig. 1.
Fig. 1.Inhibiting A. baumannii growth with B. licheniformis supernatant.
The PCR reaction created bands of 278 bp for the lanM gene and 724 bp for the lanA gene in the target probiotic strain (Fig. 2).
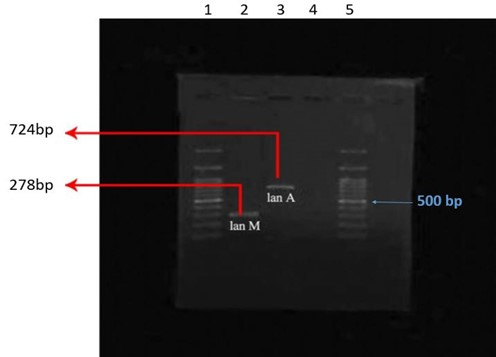 Fig. 2.
Fig. 2.Amplifying lanA, lanM genes using the PCR technique: columns 1 & 5, 100 bp ladder, column 2, lanM gene, column 3, lanA gene, column 4, negative control.
The metabolite was examined on a C18 column using the HPLC method, with the mobile phase rinsed using a blend of 1% acetic acid and acetonitrile (V/V) in a ratio of 60:40. Apart from the solvent peak (2.96 minutes with a frequency of 54.5%), the metabolite was detected at 5.96 minutes with a frequency of 6.64%, and its lipopeptide nature was determined by using the calibration curve and comparison with the Sigma international standard. The chromatogram obtained from the HPLC is shown in Fig. 3.
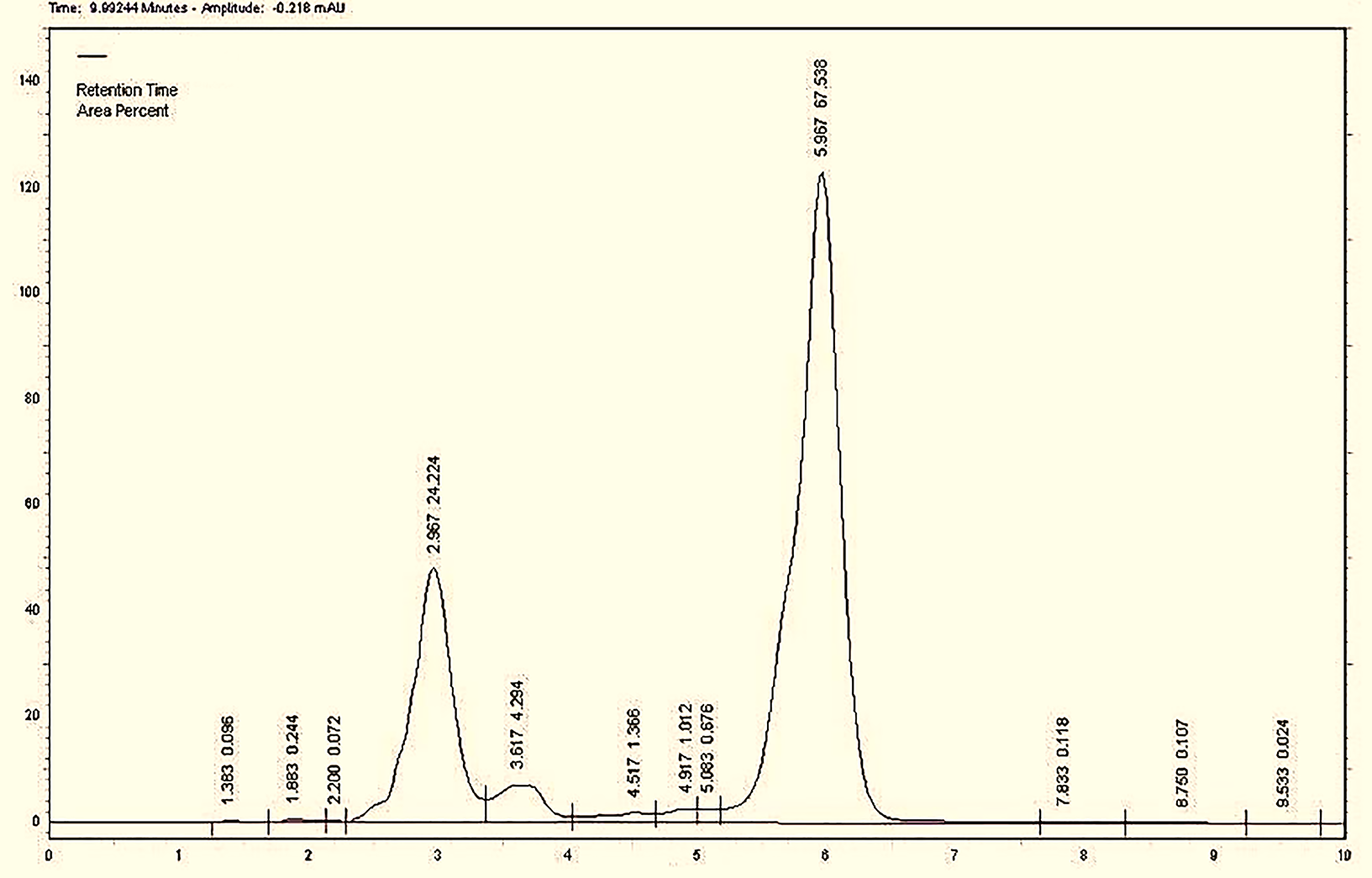 Fig. 3.
Fig. 3.Chromatogram of the metabolite obtained from the supernatant of B. licheniformis.
The chemical nature of the separated metabolites was confirmed through FTIR
spectra analysis (Fig. 4). The IR spectrum exhibited a stretching frequency of
N-H at 3300 cm
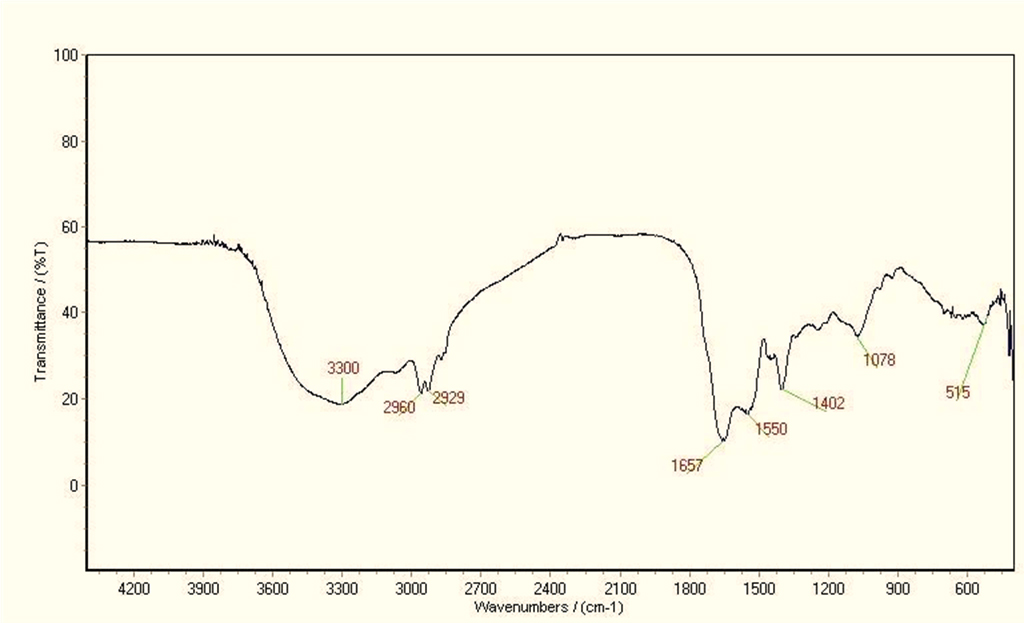 Fig. 4.
Fig. 4.Fourier-transform infrared spectroscopy (FTIR) spectrum of B. licheniformis supernatant metabolite.
Furthermore, the peak detected at 1657 cm
The Kirby-Bauer disc diffusion agar method was utilized to perform an antibiotic sensitivity test on MDR A. baumannii. In this study, these bacteria demonstrated resistance to 10 out of the 20 antibiotics that were tested. The antibiotics to which they exhibited resistance included Ceftriaxone (CRO), Ceftazidime (CAZ), gentamicin (GM), amikacin (AK), ciprofloxacin (CIP), piperacillin (PRL), piperacillin-tazobactam (PTZ), imipenem (IMI), meropenem (MEM), treonam (ATM), cefepime (CFM), Cefotaxime (CTX), ampicillin-sulbactam (SAM), tobramycin (TN), ticillin-clavulanic acid (TCC), polymyxin B (PB), Cotrimoxazole (TS), Doxycycline (DXT), Cefepime (CPM), and Levofloxacin (LEV). It should be emphasized that intermediate outcomes were also regarded as resistant [42, 51]. The detailed results can be found in Supplementary file (Supplementary Table 1).
The MIC values of the probiotic product and several ineffective antibiotics, as mentioned in the table above, were determined and compared against the specified MDR standard strain using microdilution broth. When the MIC test results were compared, it was observed that the lipopeptide derived from the probiotic exhibited a relatively low concentration, effectively suppressing the growth of drug-resistant bacterial strain. Specifically, the MIC value for the lipopeptide derived from B. licheniformis supernatant against the MDR A. baumannii ATCC BAA-747 strain was found to be 4 µg/mL. The MIC results is observable in Supplementary file (Supplementary Table 2).
Through optical absorption measurements and utilizing the aforementioned formula, it has been demonstrated that A. baumannii’s standard strain exhibits a robust biofilm formation capability prior to its proximity to the probiotic product. The OD of the sample showed an absorption value of 0.788, while the well ODC was determined to be 0.131. Following treatment with the extracted lipopeptide, the light absorbance of the sample decreased to 0.212. Based on these findings, a 73% inhibition of biofilm formation was achieved the standard strain of A. baumannii.
The FE-SEM electron microscopy images (Fig. 5a,b) at the same magnification (10.00 kx) captured from the biofilm of the MDR bacterial strain revealed the creation and inhibition of the biofilm, both prior to and following treatment with lipopeptide extracted from B. licheniformis.
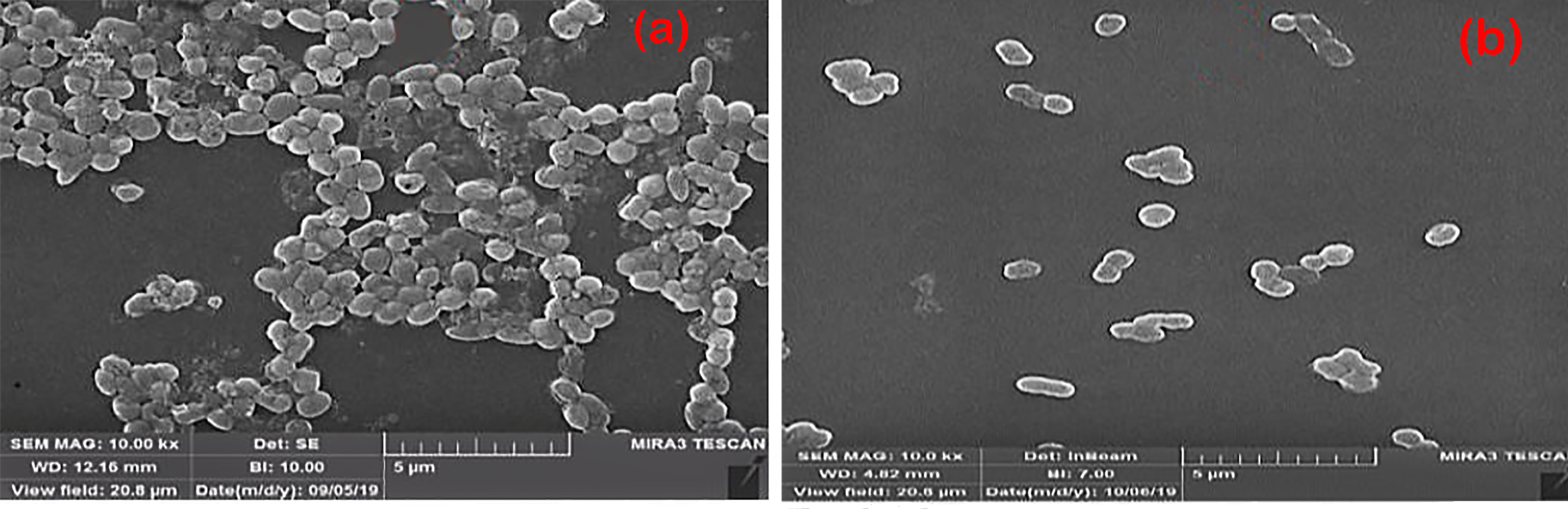 Fig. 5.
Fig. 5.Biofilm formation in A. baumannii before (a) and after (b) treatment with lipopeptide probiotic product.
The A. baumannii genome was extracted and utilized as a template for PCR amplification. Electrophoresis of the amplified products on an agarose gel revealed bands at 605 bp for the pgaA gene, 400 bp for the pgaB gene, 518 bp for the pgaC gene, and 224 bp for the pgaD gene (Fig. 6).
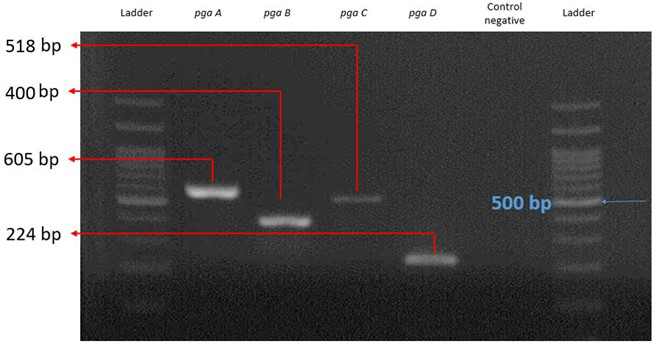 Fig. 6.
Fig. 6.Amplification of pga operon genes after gel electrophoresis next to the ladder and negative control.
The relative qPCR results were assessed prior to and following treatment a standard A. baumannii strain with lipopeptide extracted from B. licheniformis as a probiotic.
To examine and compare the expression of the mentioned genes, we require two conditions:
(A) Samples treated with different antibiotics.
(B) Samples treated with the lipopeptide extracted from B. licheniformis.
Conducting an investigation into gene expression patterns under varying
conditions, such as no treatment, treatment with one of the selected antibiotics
from Supplementary Table 2 (MEM), and treatment with a probiotic
product. To assess the expression of the target genes, the following steps were
carried out: total RNA extraction, cDNA synthesis, addition of specific primers
for each target genes (pgaA, B, C, D) and
reference gene (16sDNA), addition of Cybergreen master-mix, and
adjustment of the temperature program on the Real-time device. During the
operation of the device, the acquired data, images, graphs, and Cqs could be
reviewed and analyzed. The obtained data and
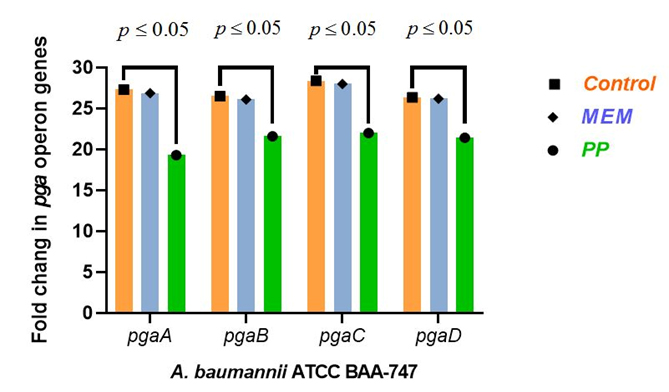 Fig. 7.
Fig. 7.Histogram of pgaA, pgaB, pgaC and, pgaD genes expression in multidrug-resistant (MDR) A. baumannii under different conditions: without treatment (Control), treatment with the selected antibiotic meropenem (MEM), and treatment with the probiotic product (PP).
Furthermore, the melting curves analysis was conducted on the target genes following the conclusion of the qPCR process. This analysis served to validate the optimal efficiency of the primers utilized in the reaction and confirmed the specific amplification of the pga operon genes (pgaA, B, C, D) in the standard strain of A. baumannii (Supplementary file (Supplementary Fig. 1)).
Drug resistance presents a significant challenge in the treatment of microbial infections, leading researchers to explore new strategies for combating resistant microbes [52]. Certain probiotic substances have been identified as having the ability to combat bacteria and assist in overcoming drug resistance [53]. In addition to their antibacterial properties, these substances offer favorable benefits for the body by promoting microbial balance and boosting the immune system [54].
Bacterial biofilms are composed of various substances, including proteins, polysaccharides, and phospholipids, which enhance bacterial resistance to antimicrobial agents. As previously mentioned, PNAG is a polysaccharide that promotes adhesion between bacteria within the biofilm and is considered a virulence factor for bacteria. The genes of the pga operon are involved in its synthesis in A. baumannii [55]. Choi et al. [16] conducted a study on the pgaABCD locus in A. baumannii and discovered that this locus codes for proteins known as PNAG, which play a crucial role in biofilm formation. They performed PCR analysis on 30 strains of A. baumannii obtained from hospitals. Out of these 30 strains, 14 exhibited robust PNAG production, 14 displayed weak PNAG production, and two strains lacked any PNAG product. By comparing two mutant strains of A. baumannii in terms of PNAG production, they observed that biofilm formation did not occur in strains lacking this particular locus [16].
In 2022, an Egyptian study examined the impact of L. rhamnosus probiotic products on Enterococcus faecalis using the gel diffusion technique. The study demonstrated the antibacterial potential of L. rhamnosus products. However, in our study, the effect of B. licheniformis probiotic products on A. baumannii was found to be more significant when compared to L. rhamnosus products [56].
Different mechanisms likely play a role in the antibiotic activity of linear and cyclic lipopeptides. This is because they are distinct amphiphiles that not only interact with the charged groups of membrane lipids through electrostatic interactions, but also with the hydrophobic regions of the lipid bilayer in membranes [57]. These interactions can result in electrostatic and mechanical changes, decreased surface tension, enhanced metal ion absorption, and disruption of lipid bilayers [58]. In simpler terms, these lipopeptides generally have a tendency to interact with membranes and form pores within them [59]. By entering the cell cytoplasm through membrane pores, these metabolites ultimately impact the transcription and expression processes of genes associated with microbial cell virulence, altering the expression of genes of interest [60]. Hence, employing advantageous microbial metabolites to minimize or hinder the activation of genes responsible for biofilm formation in A. baumannii could present a feasible approach in tackling antibiotic-resistant strains.
In our research, we have validated the lipopeptide composition of the probiotic product through the analysis of HPLC and FTIR data. Moreover, this probiotic has shown a lower MIC value against the targeted MDR bacteria when compared to other antibiotics. Electron microscopy images have illustrated the prevention of bacterial biofilm formation. Additionally, the outcomes of this research suggest that the probiotic produced by B. licheniformis influences the downregulation of pga genes, leading to the dispersion and inhibition of PNAG synthesis, subsequently resulting in defects in the bacterial biofilm.
In 2020, Porbaran et al. [61] collected 120 hospital samples from different clinical patients. The samples comprised of 50 strains of P. aeruginosa and 70 strains of A. baumannii. Similar to our study, they utilized PCR testing to validate the existence of pga operon genes and analyzed the frequency of these resistance genes. They concluded that the gene frequencies of the pgaABC operon in A. baumannii were 45.71% for pgaA, 32.85% for pgaB, and 42.85% for pgaC [61].
In their 2021 research, Gupta et al. [62] evaluated the antibiofilm potential of glycolipid obtained from B. licheniformis against Candida glabrata biofilm. They employed a scanning electron microscope (FE-SEM) to evaluate the anti-biofilm characteristics of the derived glycolipid, which inhibited and eradicated up to 80% of C. glabrata biofilm [62].
The MIC value of our probiotic product in the present study was determined to be 4 µg/mL, indicating its strong potency as an antimicrobial agent. In a 2022 study by Roberts et al. [63], investigated the antibacterial properties of a synthetic lipopeptide against multidrug-resistant Gram-negative pathogens, including A. baumannii, and MICs were reported approximately high. It is clear that the potency of the natural probiotic product derived from B. licheniformis is better than the material used in the aforementioned study.
Lipopeptides, essentially short-chain fatty acids, when consumed by bacteria,
modify bacterial metabolism. They reduce the production of adhesive substances
essential for biofilm formation while facilitating an anti-adhesion mechanism
[64, 65]. Lactobacilli employ diverse mechanisms to secrete
antimicrobial compounds, which may involve the liberation of organic acids [66].
These substances create differences in proton electrochemical gradients and
membrane permeability, resulting in increased intracellular acidity and protein
denaturation [67]. Moreover, Lactobacilli produce H
Various research have demonstrated that, in addition to probiotics, there are other compounds with antimicrobial properties. For instance, a study conducted in 2021 on the effects of cannabidiol in managing drug resistance revealed its effectiveness against highly resistant bacteria such as S. aureus and Streptococcus pneumoniae, as well its excellent activity against biofilms. Furthermore, this substance, derived from the non-psychoactive component of marijuana, possesses the ability to act locally on drug-resistant infections [69].
In 2022, another study discovered an antibacterial peptide called lycosin II, which is derived from the venom of the spider Lycosa singoriensis. Lycosin II exhibited potent antibacterial and biofilm-inhibiting properties against both Gram-positive and Gram-negative bacteria, including resistant strains like S. aureus. The study revealed that lycosin II binds to lipoteichoic acid and lipopolysaccharide present in bacterial membranes, leading to their destruction. The results indicate that lycosin II has the potential to serve as a remedy for infections induced by strains resistant to multiple drugs [70]. Unlike our research, which focused on the effects of probiotic products on resistant A. baumannii, these two studies also found that antimicrobial agents targeting other bacteria had positive effects on reducing drug resistance.
Selvaraj et al. [71] carried out research in 2020, where they utilized a natural compound called myrtenol derived from plants to inhibit genes linked to the formation of biofilm in both standard and clinical strains of A. baumannii. They effectively inhibited the activity of various target genes including bfmR, csuA/B, bap, ompA, pgaA, pgaC, and katE. Similar to our own research, their findings demonstrated a reduction in gene expression, resulting in thinner biofilms and increased vulnerability of A. baumannii strains to commonly used antibiotics like CIP and GM [71].
The current study provided evidence that probiotic products exhibit antimicrobial characteristics, rendering them effective and safe substitutes for antibiotics when combating drug-resistant bacteria. The identified lipopeptide was found to suppress the expression of genes that contribute to biofilm formation in MDR bacterial strains.
Building on these results, probiotic products produced by B. licheniformis have the potential to affect the pga operon, leading to defects in bacterial biofilms and reduced resistance. Therefore, these materials can be used to combat resistant strains of A. baumannii, providing a potential treatment option for patients with antibiotic-resistant bacterial infections. Additionally, the lipopeptide’s mechanism of action also reduces the expression of biofilm formation genes in MDR bacterial strains.
One limitation of this study is the small number of standard strains evaluated, as well as the lack of evaluation of other probiotic Bacillus products. However, considering the remarkable capacity of B. licheniformis in producing antimicrobial and anti-biofilm compounds, it is advisable to utilize them as low-risk biological agents to combat pathogenic biofilms. In the future, promising solutions for combating drug-resistant pathogenic microorganisms could be achieved through further research and advancements in probiotic applications. The rise of antibiotic resistance highlights the need for further exploration into new alternative treatments to address the growing challenge of MDR bacteria.
The findings of this study indicate that probiotics offer a promising solution in combating drug-resistant bacteria. The mechanism through which these probiotic substances operate involves targeting the genes of the pga operon and reducing their expression. This action disrupts the biofilm matrix of A. baumannii, leading to diminished bacterial solidarity and decreased resistance. Consequently, these products can be employed as a means to combat MDR bacteria, effectively eliminating resistance and mitigating the excessive use of chemical antibiotics.
The data used to support the findings of this study are available from the corresponding author upon request.
MHB and FG: Investigation and perform experiments, Writing an original draft, Formal analysis, Data curation, Writing-review & Editing; RN: Supervision, Project administration, Validation, Data curation, Writing-review & Editing; SSA: Formal analysis, Writing-review & Editing; PJ: Data curation, Software. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
No experiments were conducted on animals or humans. This research has received ethics approval with ID IR.IAU.QOM.REC.1398.004 issued by Islamic Azad University, Qom branch, Qom, Iran.
We would like to thank the Department of Microbiology, Faculty of Basic Sciences, Qom Branch, Islamic Azad University, Qom, Iran for their support.
This research received no external funding.
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
